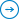Imagine going to see your doctor for chest pains, and walking out the same day with a diagnosis for a rare heart condition. That diagnosis would have taken weeks or months (at best) even a few years ago. But in this new scenario, it is now a routine outpatient test performed on-site, accompanied by an immediate treatment plan. This is a glimpse into the new face of clinical diagnosis and treatment.
Although still in the realm of imagination, such a scenario is no longer far from reality – thanks to a quiet revolution brewing in healthcare that is based on next-generation sequencing (NGS) technologies.
The first generation of DNA sequencing methods were invented in the late 1970s – the advent of the dideoxy Sanger method – which is still the gold standard for most genetic testing around the world. Over the next two decades, gene sequencing steadily advanced from a laborious process into an industrialized toolkit. As you can see from Figure 1, by 2003 the creation of innovation assets peaked in the first wave of gene sequencing technologies, with more than 5,000 patents filed or granted.
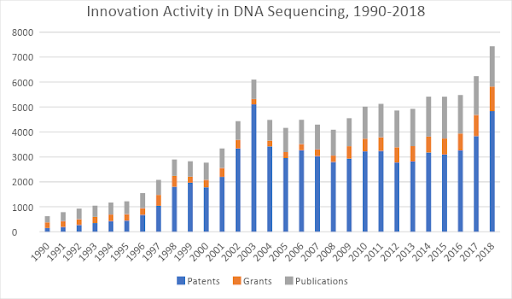 Figure 1: Patents, research grants, and scientific journal publications related to DNA sequencing, 1990-2018. Source: Wellspring search data.
Figure 1: Patents, research grants, and scientific journal publications related to DNA sequencing, 1990-2018. Source: Wellspring search data.
But that was only the beginning. First-generation gene sequencing was useful primarily for academic studies and scientific research. By the early 2000s, when the Human Genome Project was in full swing, it took 15 months and $300 million dollars just to generate a “draft” human genome sequence. The technology was still far too slow and too expensive for widespread use in the healthcare industry. As a result, only straightforward genetic lab tests for single-gene conditions were possible in a clinical setting. Unfortunately, most human diseases and disorders are caused by a multiplicity of genes or are caused by pathogens for which the risks are heightened or reduced by the presence or absence of a wide range of genes. As such, genetic testing has remained beyond reach in all but a small subset of clinical situations.
Enter Next-Generation Sequencing (NGS). Development of this second wave of technologies was underway by 2005, although the term NGS didn’t appear in earnest until 2012. In essence, NGS techniques allow for massive parallel sequencing of small slices of a genome, effectively allowing for large-scale sequencing almost in real-time. By 2018, these advances had taken the cost of sequencing a human genome down to as low as $200 and the time required to only one hour.
The result: NGS techniques have become increasingly ubiquitous, not just in scientific research but also in clinical lab testing and diagnostics. Complex medical conditions arising from many interrelated genetic factors are now diagnosable in the clinic thanks to NGS – ranging from kidney disease to heart conditions to blindness and deafness to genetically inherited forms of cancer.
Even still, this is just the beginning of what’s possible. To unlock the broad wave of disruptive effects waiting on the horizon, another wave of innovations is needed in order to make high-throughput genetic screening an accepted, ubiquitous “platform technology” in clinical practice. And indeed, a third wave of technology development is already underway. Figure 2 shows its early growth – starting in 2012 and 2013 and beginning to gain steam more rapidly today.
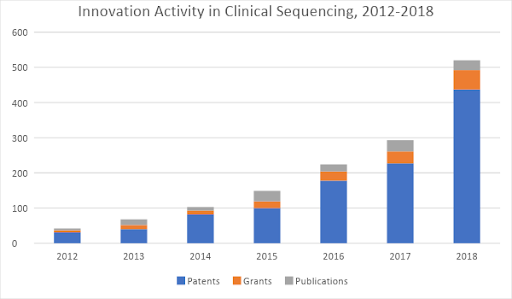
Figure 2: Patents, research grants, and scientific journal publications related to clinical applications of next-gen sequencing, 2012-2018. Source: Wellspring search data.
The promise for disruptive change exists on multiple dimensions. Imagine if screening patient genomes becomes a routine practice and if enough patients suffering from certain conditions have been sequenced. Under these conditions, we should be able to rapidly advance the understanding of which genes are implicated in a wide variety of diseases that still confound the medical profession today. If real-time sequencing technologies are cheap and portable enough, then the use of sequencing data in clinical bacteriology might allow doctors in the field to be able to, for instance, detect and stop the spread of an Ebola outbreak before it gets going. The applications are seemingly endless, and the possibilities are staggering.
To enable such a revolution in healthcare practices, a variety of enabling technologies must come together. There is already initial promise in a “third generation” of sequencing approaches – headlined by the so-called “single-molecule sequencing” method – which may lead to significant improvements in the portability and speed of sequencing compared to what is possible today.
Also needed are major improvements in bioinformatics software and analytics, to help medical professionals analyze the vast amounts of data to provide in-context diagnostic insights relevant to a given patient’s case. Companies such as Edico Genome and Seven Bridges are among a growing vanguard of new startups making headway in this space.
Over the past few decades, the advances of gene sequencing and genomic analysis have been of primary interest to companies that produce or use sequencing machines. Going forward, as the revolution in genetics and genomics makes its way into everyday clinical practice, the ripple effects will be felt by a much wider set of industries – ranging from health insurance providers to medical devices companies and everyone in between.